Research
With the rapid advancement of modern technology, data containing spatial information is being generated in increasingly larger volumes and more complex structures, presenting significant statistical challenges to the scientific community. However, I believe that rather than being a burden, the spatial nature represents a valuable opportunity, imbuing datasets with elegance and smoothness. By employing appropriate formulations, we can unlock the full potential of spatial knowledge and extract unique, highly valuable insights from the data. My research has been motivated by the statistical challenges at the forefront of science across various different spatial contexts, which require novel and efficient methodology.
Neural Networks for Geospatial Data
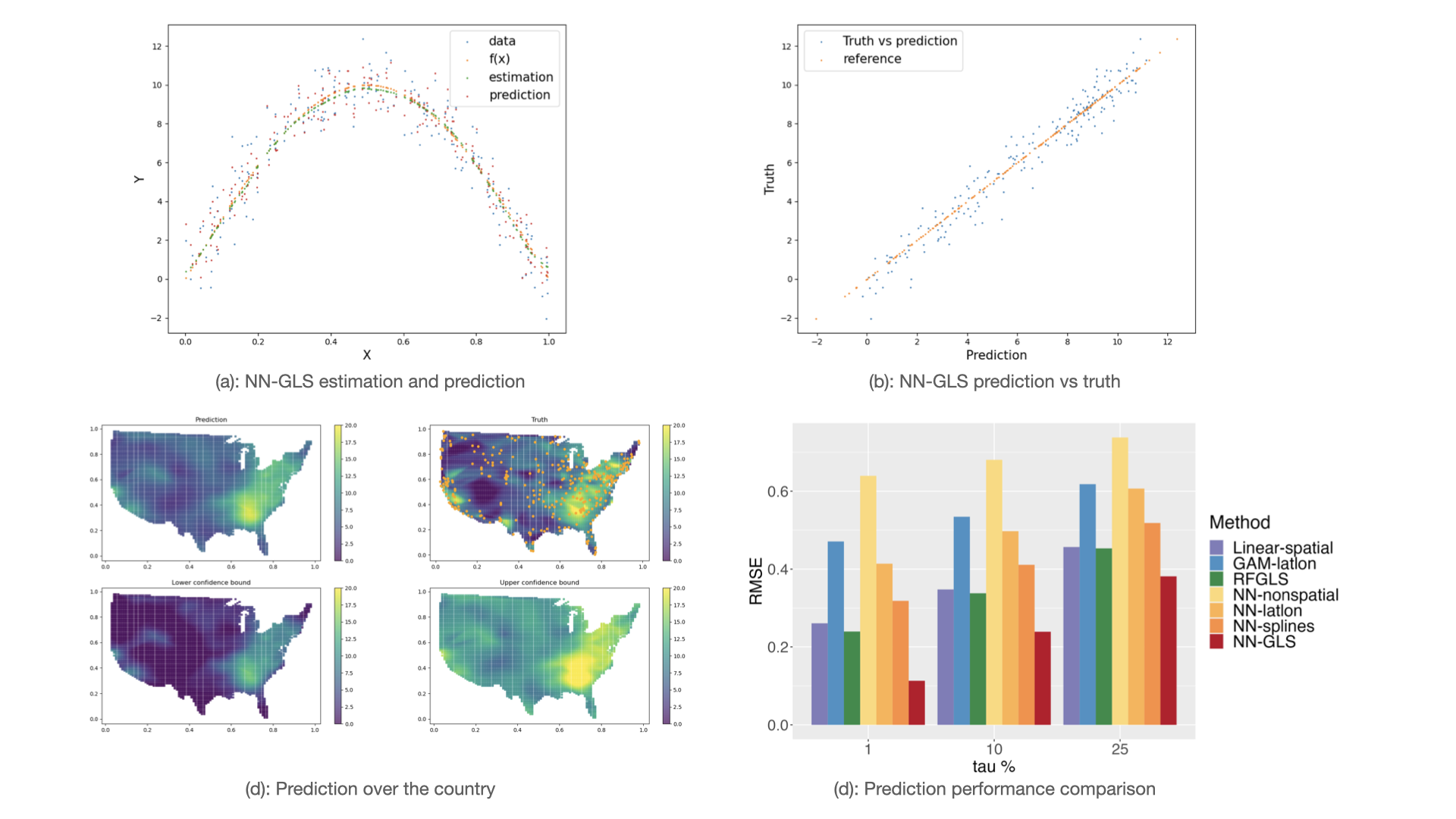
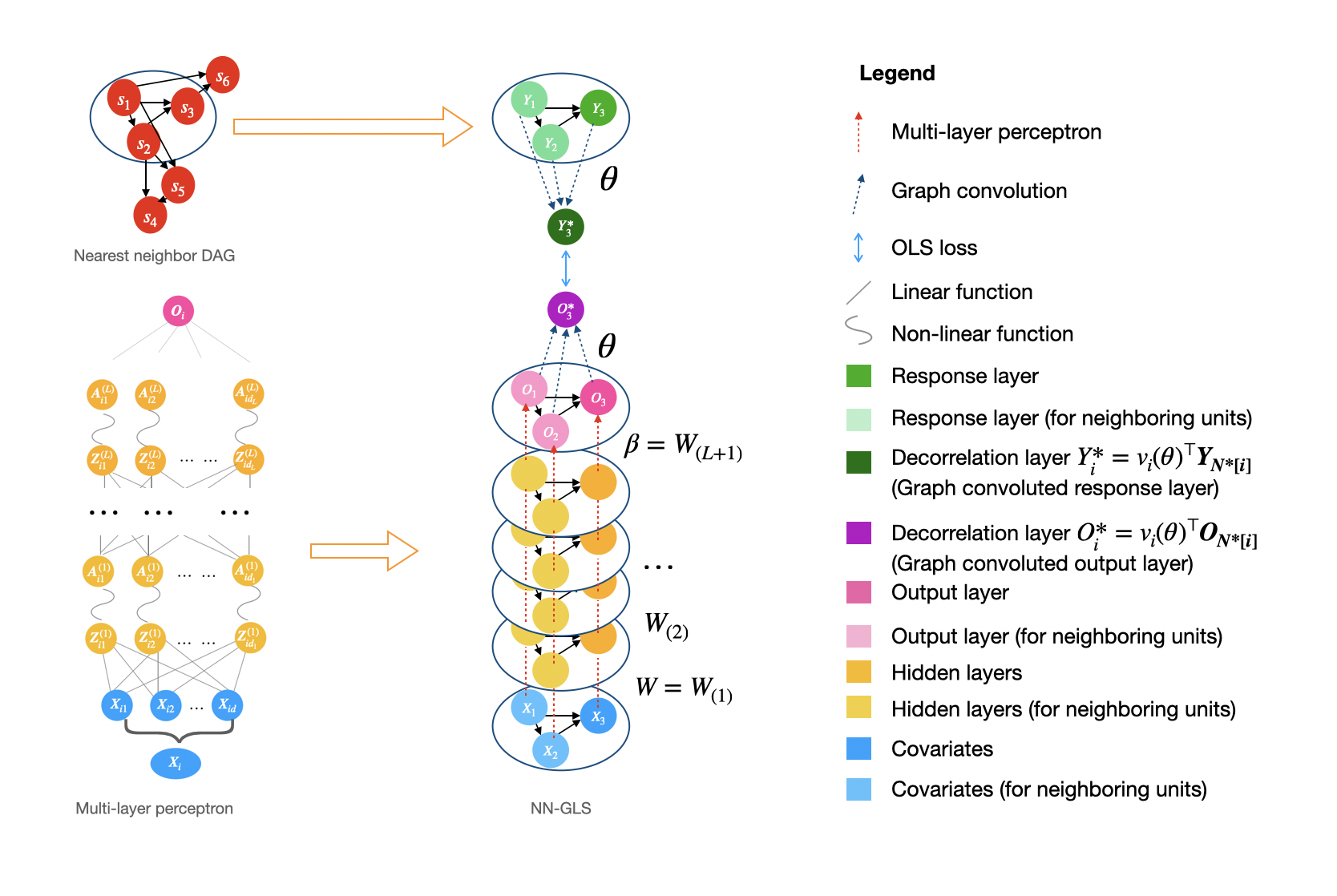
Analysis of geospatial data has traditionally been model-based, with a fixed mean model, customarily specified as a linear regression on the covariates, and a Gaussian process covariance model, encoding the spatial dependence. We propose NN-GLS that embeds neural networks directly within the traditional Gaussian process (GP) geostatistical model to accommodate non-linear mean functions while retaining all other advantages of GP, like explicit modeling of the spatial covariance and predicting at new locations via kriging.
We provide the first large-sample results for any neural network algorithm for irregular spatial data, including the consistency and finite sample concentration rates which quantifies the need to accurately model the spatial covariance in neural networks for dependent data.
NN-GLS admits a representation as a special type of graph neural network, which takes kriging weights for graph convolution. The idea can be easily generalized to a wider range of deep-learning approach, which led to “spatially-informed deep-learning” as a promising future direction.
Related publications
- Zhan, W., & Datta, A. (2024). Neural networks for geospatial data. Journal of the American Statistical Association 120 (549), 535-547. link
GeospaNN: A python implementation for NN-GLS
GeospaNN is a package for geospatial analysis using neural networks that explicitly accounts for spatial correlation in the data. The package implements the NN-GLS method and is developed using PyTorch and under the framework of PyG library. GeospaNN is a geographically-informed Graph Neural Network (GNN) for analyzing large and irregular geospatial data, that combines multi-layer perceptrons, Gaussian processes, and generalized least squares (GLS) loss. GeospaNN offers both regression function estimation and spatial prediction. The sparse approximation in NN-GLS allows efficient computation in geospaNN, which scale up to sample sizes of hundreds of thousands.
Related publications
- Zhan, W., & Datta, A. (2025+). GeospaNN: a Python package for geospatial neural networks (Under revision of Journal of Open Source Software)
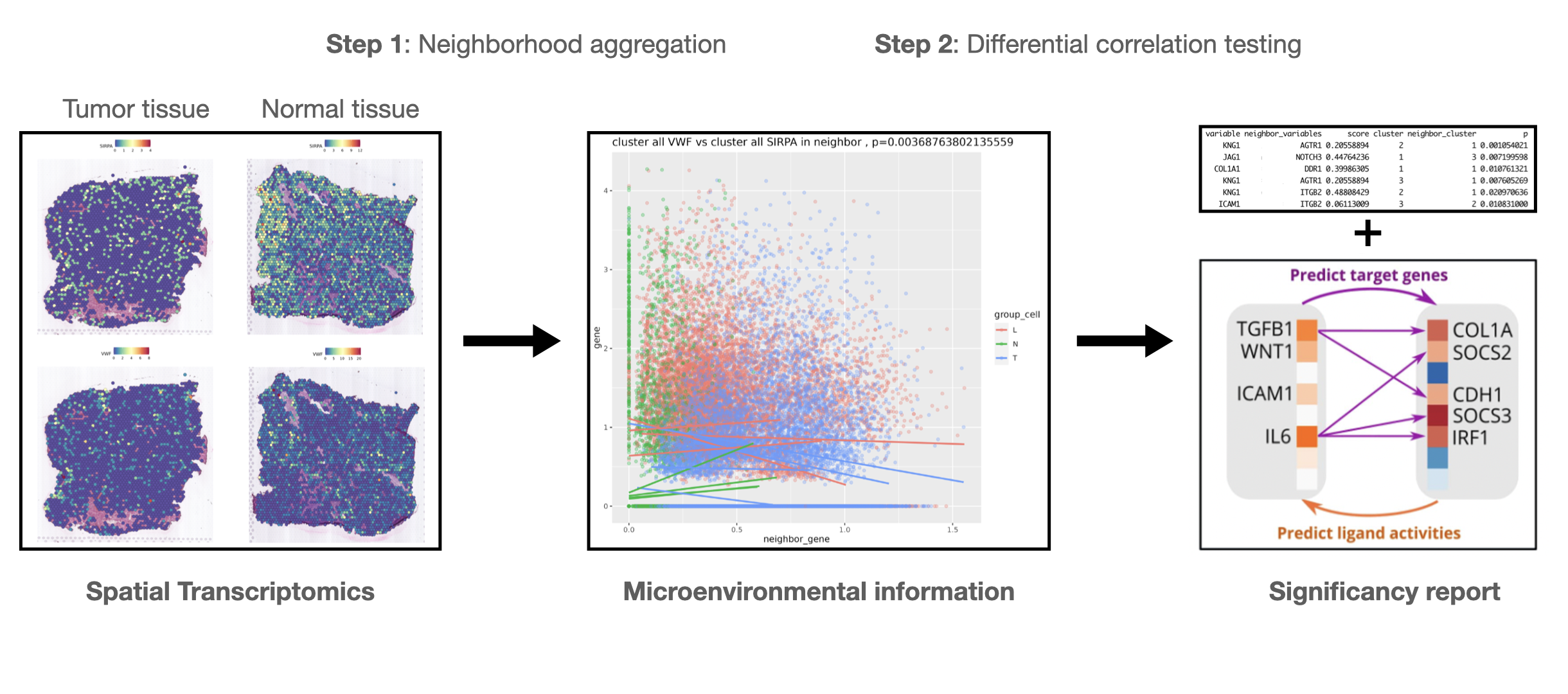
Multispa: a multi-sample cell-microenvironment analysis tool
The development of spatial transcriptomics allows for single-cell (or near-single-cell) level sequencing of tissues while preserving spatial information. This technique represents an unprecedented advancement in medical and biological research. For instance, in immunology, immune cells interact with tumor cells through antibody signaling, and the fate of a tumor cell is influenced by the density of immune cells in its microenvironment, which can only depicted by introducing “spatial distance.”
In this project, we hypothesize that the relationship between the expression of gene A in tumor cells and the expression of gene B in immune cells within their microenvironment may differ across patient groups (e.g., treatment vs. control).
We introduce Multispa, a statistical tool that leverages spatial information to identify differentially associated gene pairs between groups. Multispa uncovers distinct gene regulatory mechanisms across these groups and holds significant implications for immunotherapy research.
Related publications
- Zhan, W., & Ji, H (2025+). Multispa: a Multi-sample Cell-microenvironment Tool for Spatial Transcriptomics (Manuscript in prepartion)